_______________________________________________________________________________________________
R is an open-source statistical programming language. It is used to manipulate data, to perform statistical analyses, and to present graphical and other results. R consists of a core language, additional ‘packages’ distributed with the R language, and a very large number of packages contributed by the broader community. Packages add specific functionality to an R installation. R has become the primary language of academic statistical analyses, and is widely used in diverse areas of research, government, and industry.
R has several unique features. It has a surprisingly ‘old school’ interface: users type commands into a console; scripts in plain text represent work flows; tools other than R are used for editing and other tasks. R is a flexible programming language, so while one person might use functions provided by R to accomplish advanced analytic tasks, another might implement their own functions for novel data types.
As a programming language, R adopts syntax and grammar that differ from many other languages: objects in R are ‘vectors’, and functions are ‘vectorized’ to operate on all elements of the object; R objects have ‘copy on change’ and ‘pass by value’ semantics, reducing unexpected consequences for users at the expense of less efficient memory use; common paradigms in other languages, such as the ‘for’ loop, are encountered much less commonly in R.
Many authors contribute to R, so there can be a frustrating inconsistency of documentation and interface. R grew up in the academic community, so authors have not shied away from trying new approaches. Common statistical analyses are very well-developed.
‘Base’ R provides:
Additional packages provide:
_______________________________________________________________________________________________
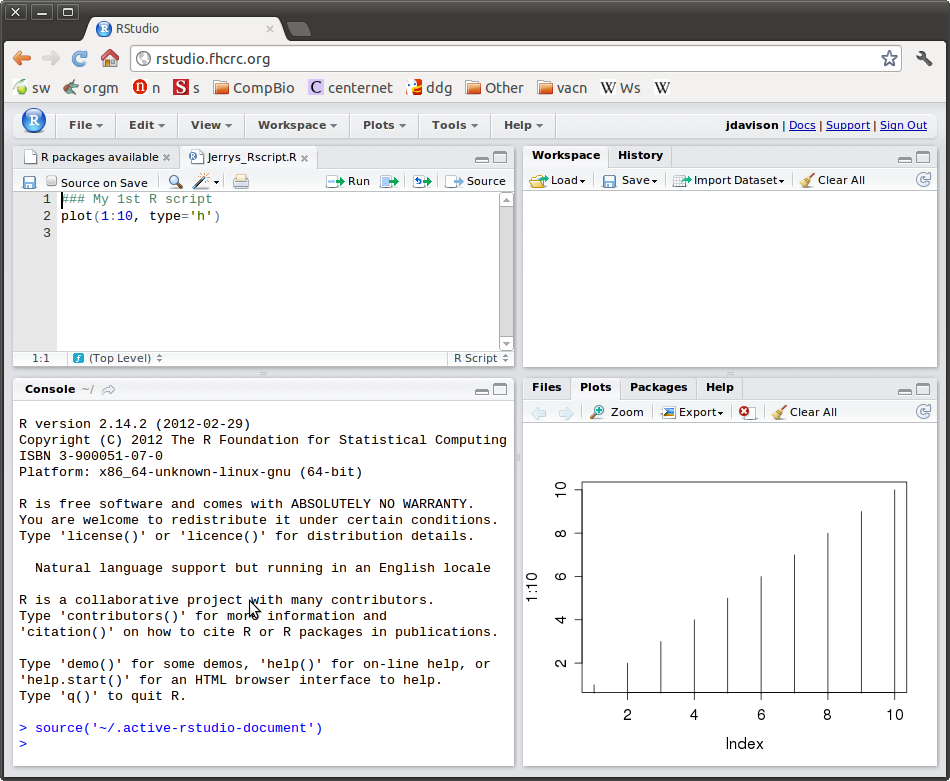
The RStudio application provides a convenient and flexible environment for your work with R. Figure 1 presents a view of a typical RStudio session.
See http://www.rstudio.com/ide for documentation and downloads for offline work on Linux, Windows and MAC systems.
R comes with extensive help. In RStudio, click the ‘help’ menu and choose ‘R help’. Important sections include
When not using RStudio, start the help system by typing the command help.start(). The R web site provides many useful links. Once you are comfortable with R, the R-help mailing list can be a very useful source of information, as can general-purpose forums like StackOverflow.
_______________________________________________________________________________________________
R is a quite sophisticated system for data analysis, but that doesn’t mean it’s not comprehensible to beginners. Let’s start using it:
∙ One supported data type is the vector, specified in this way:
∙ Data objects can be given names in an assignment statement:
∙ Exercise: evaluate these expressions: y/x, x-2/10, (x-2)/10
_______________________________________________________________________________________________
Spreadsheet applications and R complement each other in that the former can provide nicely formatted columns, colored headers and convenient scrolling while R provides functional flexibility. You can access Excel worksheets and other table-like data files with R, and use R to write files readable by spreadsheets – R reads and writes tables written in comma- or tab-separated values formats.
We’ll first read the table ALLannotationFromExcel.txt that contains ALL (acute lymphoblastic leukemia) patient information:
∙ Then use R functions that tell us about the file:
∙ Exercise: Read file ALLmetadata.txt from the same directory as before, assigning the name ’doc’ to the data frame created. How does ’doc’ relate to ’info’?
_______________________________________________________________________________________________
R doesn’t provide scrollbars like spreadsheet applications do, but you can examine subsets of large objects like the 127x21 info data frame – for example by explicitly giving the rows and columns you want to see:
∙ First and last rows of data frames:
∙ R help is handy. For example – does "head" always presents 6 rows?
∙Exercise: List the first 10 rows of columns ’sex’, ’remission’ and ’date.last.seen’ in data frame info.
∙ Data frame column names can be used to access their values:
∙ You can subset using logical expressions – watch out for NA’s!
_______________________________________________________________________________________________
R provides several data types that follow rules you might expect: the numeric can be used in arithmetic operations, the logical results from TRUE or FALSE questions, character data are used to handle text, and factors are used in statistical analyses, ANOVA for example.
∙Exercise: Use the table function to count the number of ’M’ and ’F’ patients identified in the info data frame. Does that account for all patients?
_______________________________________________________________________________________________
Select a subset of patients with normal cytogenetics and no translocations to write to a separate file and then read with a spreadsheet application: